Introduction
Ontologies and controlled vocabulary
An ontology is a formal and explicit specification of concepts and their relationships in a particular domain. It defines a shared vocabulary for that domain, including terms and their definitions, and the relationships between those terms. Ontologies are used to represent knowledge in a way that can be processed by machines, making it easier to integrate and use information across different systems and applications.
Controlled vocabularies, on the other hand, are lists of standardized terms or phrases that are used to describe a particular domain. These vocabularies are often used to index and retrieve information, ensuring that data is consistent and can be found using standardized terminology.
The importance of ontologies and controlled vocabularies for semantic text annotations lies in their ability to provide a common language for communication between different systems and applications. By using a shared vocabulary, different users and applications can understand each other's data, making it easier to share and integrate information across different platforms.
In addition, ontologies and controlled vocabularies can help to improve the accuracy and completeness of annotations. By using standardized terms and definitions, annotators can ensure that their annotations are consistent and that they accurately reflect the content of the text.
Not using ontologies for text annotation in the scientific domain can have several downsides.
Without ontologies, there may be a lack of standardized and structured terminology, leading to inconsistencies and ambiguity in annotations. This can make it challenging to integrate and compare data from different sources or perform accurate data analysis.
The absence of ontologies hinders interoperability and data sharing, as there is no common framework for understanding and linking concepts across different scientific domains.
Without ontologies, it becomes difficult to leverage existing knowledge bases and resources, limiting the potential for automated reasoning, knowledge discovery, and advanced NER training.
Overall, not using ontologies in scientific text annotation can impede collaboration, hinder knowledge integration, and limit the overall scientific understanding and advancements in the field.
The absence of ontology annotations in scientific data capture, annotation, and deposition can be attributed to resource-intensive ontology development and maintenance. Constructing ontologies that accurately represent the complexities of scientific concepts and relationships requires substantial domain expertise, time, and effort. This can be particularly challenging due to the rapidly evolving nature of scientific knowledge. The associated costs and efforts involved in ontology development often discourage widespread adoption in the scientific domain. However, initiatives like NFDI4Chem - TIB Terminology Service and EMBL - EBI OLS4 provide valuable resources for querying and retrieving ontology terms, helping to address this challenge.
Consequently, the lack of ontology-driven rich input fields in web applications and Electronic Lab Notebooks (ELNs) poses another issue. This absence limits the ability to incorporate structured ontology-based annotations directly within forms, hindering the capture of semantically meaningful data. The integration of ontology-driven input fields would enable researchers to capture and annotate data in a manner aligned with domain-specific ontologies, facilitating improved data quality, interoperability, and knowledge integration within the scientific community.
Webcomponents
Web components are a set of standardized web platform APIs (Application Programming Interfaces) for creating reusable, encapsulated, and interoperable custom HTML elements. They allow developers to define their own HTML tags and elements with associated JavaScript logic, styles, and templates, which can then be used across multiple web pages and applications. Web components make it easier to build complex web applications by promoting modularity, reusability, and extensibility, and can improve the performance, maintainability, and accessibility of web content.
The primary benefit of custom html elements is that they can be used with any framework, or even without a framework. This makes them ideal for distributing components where the end consumer may not be using the same frontend stack, or when you want to insulate the end application from the implementation details of the components it uses.
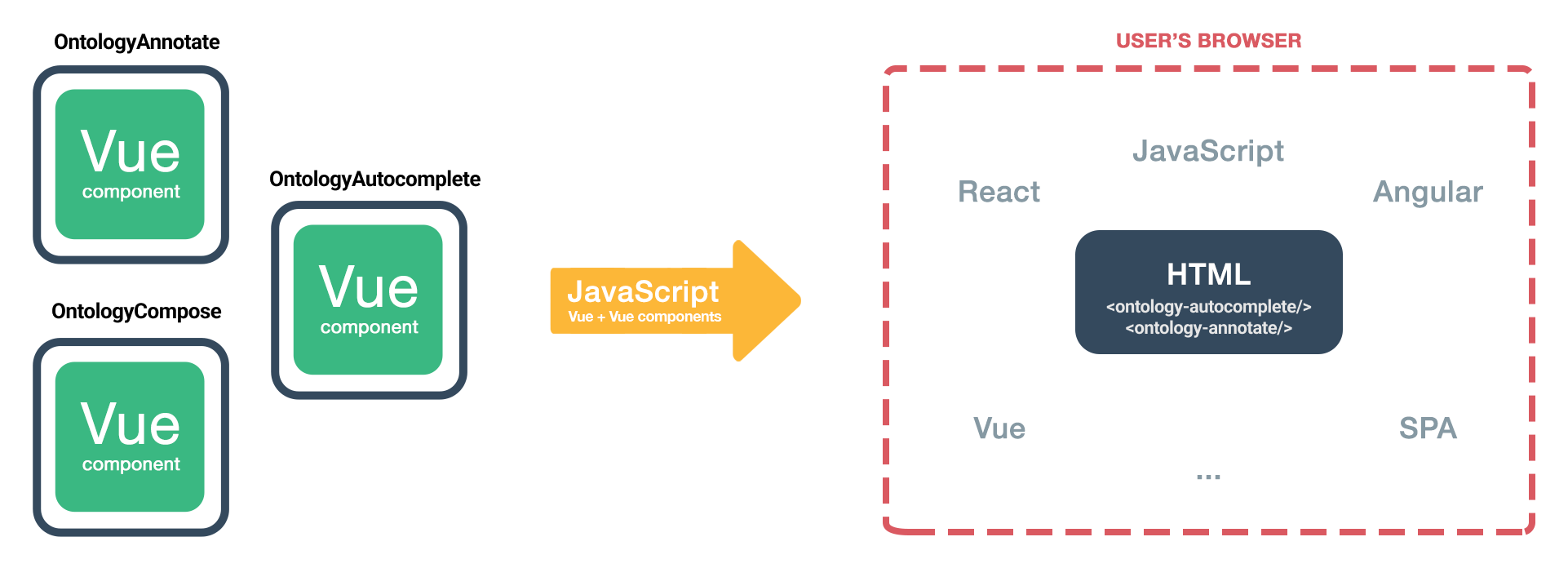
Ontology-elements
Ontology-Elements project aims at developing custom ontology rich html elements (web components).These rich ontology driven HTML elements can play a significant role in improving text annotation by providing a structured and semantically rich representation of the content. Here's how ontology-driven HTML elements can help in text annotation.
Semantic Markup: By utilizing ontology-driven HTML elements, text can be marked up with semantic tags that represent specific concepts or entities within the text. These semantic tags can capture the meaning and relationships of different elements, making it easier to interpret and annotate the content accurately.
Enforcing Consistency: Ontology-driven HTML elements ensure that the annotations adhere to a predefined ontology, enforcing consistency in the annotations across different texts. This consistency helps in organizing and retrieving annotated data effectively.
Interoperability and Integration: Ontology-driven HTML elements allow for easy integration of annotated text with other systems and applications that understand the underlying ontology. This interoperability facilitates data sharing, data integration, and the ability to leverage existing tools and resources that work with the ontology.
Richer Annotation: Ontologies provide a comprehensive representation of domain knowledge, including hierarchical relationships, attributes, and constraints. By using ontology-driven HTML elements, annotations can capture not just the surface-level information but also the contextual relationships and additional attributes associated with the annotated text.
Improved Search and Retrieval: Ontology-driven annotations enable more sophisticated search and retrieval capabilities. Users can query the annotated text based on the concepts and relationships defined in the ontology, allowing for more precise and targeted retrieval of information.
Domain-Specific Annotation: Ontology-driven HTML elements can be tailored to specific domains or application requirements. This customization allows for domain-specific annotations that capture the nuances and specificities of the given domain, leading to more accurate and relevant annotations.
Text Markup
One of the primary goals of the ontology-elements project is to reuse existing components (formats, tools etc.) in the industry and be able to capture, persist and retrieve data when necessary. Different life sciences projects developed or adapted different formats to store the ontology annotations.
For example:
UniProt
Here's an example of a UniProt Gene Ontology (GO) annotation and how it is typically stored in the UniProt database:
UniProt accession: P12345
UniProt entry name: PROTEIN_A
Gene Ontology annotation:
GO term: GO:0003677
GO term name: DNA binding
Evidence code: IEA (Inferred from Electronic Annotation)In the UniProt database, the GO annotations are stored in a structured format called the Gene Ontology Annotation (GOA) file. Each annotation consists of several fields that capture relevant information.
Here's an example of how the GO annotation for the UniProt entry "PROTEIN_A" with accession number "P12345" would be stored in the GOA file:
UniProtKB P12345 PROTEIN_A GO:0003677 IEAIn this example, the fields represent the following information:
UniProtKB: The UniProt Knowledgebase identifier (accession number) for the protein.
P12345: The UniProt entry name for the protein.
PROTEIN_A: The protein's official name or description.
GO:0003677: The GO term identifier for the DNA binding term.
IEA: The evidence code indicating that the annotation was inferred from electronic annotation.
The GOA file contains entries for multiple proteins, and each annotation follows the same format, allowing efficient storage and retrieval of GO annotations. These annotations can be accessed through the UniProt website or downloaded for further analysis.
ClinicalTrials.gov
ClinicalTrials.gov does not directly use a disease ontology for its annotations. However, it does utilize the Medical Subject Headings (MeSH) controlled vocabulary, which includes a broad range of terms related to diseases, conditions, and medical concepts. MeSH terms are used to annotate clinical trials in the database. Here's an example of a ClinicalTrials.gov annotation using MeSH terms and how it is stored in the database:
ClinicalTrials.gov Identifier: NCT01234567
Title: Study of Drug X in Patients with Hypertension
MeSH Annotation:
MeSH Term: Hypertension
MeSH Unique Identifier (UI): D006973In this example, the fields represent the following information:
ClinicalTrialID: The unique identifier assigned to the clinical trial in ClinicalTrials.gov.
Title: The title of the clinical trial.
MeSHAnnotations: A collection of MeSH annotations associated with the clinical trial.
MeSHTerm: The MeSH term representing the disease or medical concept.
MeSHUI: The unique identifier (UI) assigned to the MeSH term in the MeSH controlled vocabulary.
These annotations help categorize and index clinical trials based on their relevant diseases or medical concepts, enabling researchers and users to search and retrieve trials related to specific conditions.
FlyBase
FlyBase, a database dedicated to the fruit fly Drosophila melanogaster, utilizes ontologies such as the FlyBase Controlled Vocabulary (FBcv) and the FlyBase Anatomy Ontology to annotate genes, phenotypes, and anatomical structures. Here's an example of a FlyBase ontology annotation and how it may be stored in the database:
FlyBase Gene ID: FBgn0012345
Gene Symbol: GeneA
Annotation:
Annotation Type: Phenotype
Term: FBcv:0001234
Term Name: Abnormal wing morphology
Evidence: IMP (Inferred from Mutant Phenotype)In the FlyBase database, ontology annotations are typically stored in structured formats, such as tables or documents, that capture the relevant information. Here's an example of how the FlyBase ontology annotation for the gene "GeneA" with FlyBase Gene ID "FBgn0012345" and the phenotype "Abnormal wing morphology" would be stored:
FlyBase Gene ID Gene Symbol Annotation Type Term Term Name Evidence
FBgn0012345 GeneA Phenotype FBcv:0001234 Abnormal wing morphology IMPIn this example, the fields represent the following information:
FlyBase Gene ID: The unique identifier assigned to the gene in FlyBase. Gene Symbol: The symbol or name associated with the gene. Annotation Type: Specifies the type of annotation, such as "Phenotype" in this case. Term: The identifier or code representing the ontology term associated with the annotation. Term Name: The name or description of the ontology term. Evidence: Indicates the evidence code supporting the annotation, such as "IMP" (Inferred from Mutant Phenotype) in this example.
By storing the FlyBase ontology annotations in a structured format, the database allows for efficient retrieval, querying, and analysis of gene and phenotype data based on the associated ontology terms, providing researchers with valuable information for studying the genetics and biology of Drosophila melanogaster.
MetaboLights
MetaboLights stores its ontology annotations using the ISA-Tab format. The ISA-Tab format is a widely adopted standard for organizing and describing metadata associated with biological experiments, including metabolomics studies.
In the context of MetaboLights, the ISA-Tab format provides a structured and standardized framework for capturing and storing ontology annotations alongside experimental metadata. The annotations are typically stored within the metadata files associated with each study or assay. The ISA-Tab format organizes metadata into three main components: investigation, study, and assay. Within these components, ontology annotations are stored using specific columns or fields dedicated to capturing ontology terms. Researchers can associate relevant ontology terms with various aspects of their experiments, such as sample characteristics, experimental variables, or data annotations.
Here is an example MetaboLights samples ontology annotation
Source Name Characteristics[Organism] Term Source REF Term Accession Number Characteristics[Organism part] Term Source REF Term Accession Number Protocol REF Sample Name Factor Value[Gender] Term Source REF Term Accession Number Factor Value[Metabolic syndrome] Term Source REF Term Accession Number
ADG10003u Homo sapiens NCBITAXON http://purl.obolibrary.org/obo/NCBITaxon_9606 urine BTO http://purl.obolibrary.org/obo/BTO_0001419 Sample collection ADG10003u_007 Male diabetes mellitus
ADG10003u Homo sapiens NCBITAXON http://purl.obolibrary.org/obo/NCBITaxon_9606 urine BTO http://purl.obolibrary.org/obo/BTO_0001419 Sample collection ADG10003u_008 Male diabetes mellitus
ADG10003u Homo sapiens NCBITAXON http://purl.obolibrary.org/obo/NCBITaxon_9606 urine BTO http://purl.obolibrary.org/obo/BTO_0001419 Sample collection ADG10003u_009 Male diabetes mellitus
ADG10003u Homo sapiens NCBITAXON http://purl.obolibrary.org/obo/NCBITaxon_9606 urine BTO http://purl.obolibrary.org/obo/BTO_0001419 Sample collection ADG10003u_010 Male diabetes mellitusStandoff format
All the above examples are developed and adapted to fit the requirements of particular database of interest. But project like ontology-elements would need a generic format to store ontology annotations (including its classes) and relationships etc.
This is where the standoff format comes into picture.
The standoff format, also known as the annotation standoff format, is a method for representing annotations or markup in text documents independently from the document's content. It involves storing the annotations in a separate file or data structure, associating them with specific spans or positions within the document.
BRAT (Brat Rapid Annotation Tool) is a popular open-source tool used for manual annotation of text documents. The BRAT standoff format is a specific implementation of the standoff format used by the BRAT tool to represent annotations.
This provides all the necessary features for text annotation such as separation of concerns, flexibility, compatibility, reusability and preservation.
Here is an example of a Standoff format
# text:
The quick brown fox jumps over the lazy dog.
Standoff format annotations:
T1 Animal 16 19 fox
T2 Animal 40 43 dogOntology annotation - Standoff format
Ontology elements project stores the ontology driven annotations in the standoff format but deviates in a few ways. To name a few the text and its annoations are stored in the same document or database field to avoid schema changes (primary requirement to facilitate easy adoption). It also extends the standoff annotations structure to facilitate storage of ontology IRI and classes.
Here is an example.
# text
Seven previously undescribed diterpenoids, tinocrisposides A–D (1–4) and borapetic acids A (5), B (6), and C (7), together with 16 known compounds, were isolated from the stem of Tinospora crispa (Menispermaceae). The structures of the new isolates were elucidated by spectroscopic and chemical methods. The β-cell protective effect of the tested compounds was examined on insulin-secreting BRIN-BD11 cells under dexamethasone treatment. Diterpene glycosides 12, 14–16, and 18 presented a substantial protective effect on BRIN-BD11 cells treated with dexamethasone in a dose-dependent manner. Compounds 4 and 17 with two sugar moieties exhibited clear protective effects on β-cells.# annotation
Seven previously undescribed diterpenoids, tinocrisposides A–D (1–4) and borapetic acids A (5), B (6), and C (7), together with 16 known compounds, were isolated from the stem of Tinospora crispa (Menispermaceae). The structures of the new isolates were elucidated by spectroscopic and chemical methods. The β-cell protective effect of the tested compounds was examined on insulin-secreting BRIN-BD11 cells under dexamethasone treatment. Diterpene glycosides 12, 14–16, and 18 presented a substantial protective effect on BRIN-BD11 cells treated with dexamethasone in a dose-dependent manner. Compounds 4 and 17 with two sugar moieties exhibited clear protective effects on β-cells.
$$$$
c CHEBI 29 41 diterpenoid http://purl.obolibrary.org/obo/CHEBI_23849
c SNOMEDCT 197 211 Family Menispermaceae (organism) http://snomed.info/id/107532005
c CHEBI 413 426 dexamethasone http://purl.obolibrary.org/obo/CHEBI_41879
c CHEBI 438 461 diterpene glycoside http://purl.obolibrary.org/obo/CHEBI_71939
c CHEBI 551 564 dexamethasone http://purl.obolibrary.org/obo/CHEBI_41879