Workshop Overview
Date and Location
Jena: 19.07.2023
Participants (Alphabetically)
Prof. Christoph Steinbeck, Dr. Eva Ziegler, Dr. Johannes Liermann, prof Luc Patiny, Dr. Nils Schlörer , Ms. Nisha Sharma, Ms. Noura Rayya, Dr. Pavel Kessler, Dr. Stefan Kuhn, Dr. Steffen Neumann, Dr. Tillmann Fischer, Mr. Venkata Chandra Sekhar Nainala
Recommendations Standard for Reporting Liquid-State NMR Experiments of Small Molecules
This recommendations standard is a proposal from NFDI4Chem of what metadata to report for liquid-state NMR experiments of small molecules. We aim to make it suitable for NMR repositories and the supplementary materials of publications, not for the metadata in the experiment section. Therefore, we want to emphasize that the chemist doesn't need to enter their data in the recommended format as long as the repository is capable of converting it. For instance, the user might only upload NMR datasets, then NMRium extracts the solvent, and the repository does the mapping to the CHEBI instance.
While forging this standard, we have considered two aspects: How to make sense of the data on the first hand, where we need the minimum mentioned here in the first level, and data traceability and reproducibility, where details like the number of scans and the instrument become very useful, so we asked for them in the second level.
The recommendations started with metadata for reporting NMR analyses of synthetic compounds only, covering the reaction leading to the analyzed product. Then the decision was taken to split the recommendations and remove the synthetic compounds section to make them more modular and broader to cover all small molecules, including natural products, synthetic compounds, and mixtures where we have known molecules. They also started by making crucial and automatically extractable metadata (by NMRium) mandatory, which can encourage chemists to use automatic workflows, and we made metadata relevant to impurities recommended. However, for historical data, authors might fail to submit with restricted minimum information (MI), which led to the decision to keep automatically extractable metadata recommended if it wasn't crucial.
Here is the link to the recommendations draft, which will be open for editing until publication.
Scientific Applications Development - NMRium
Here we show Zakodium approach for the development of NMRium and how to make programming fair. Central to their methodology is the creation of highly sought-after libraries, such as React components including parsers, jcamp or Bruker converters, and tools for generating jcamp from other data.Zakodium adopts the practice of conventional committing to facilitate the automated generation of the change log based on bug fixes and new features, seamlessly closing associated issues. They harness continuous integration and leverage automatic GitHub testing. "Coverage" tells how much of the code was tested and which lines weren't, usually over 80% of the code is tested.
To ensure the visibility and accessibility of their projects, Zakodium utilizes the Zenodo platform for publication, thereby obtaining Digital Object Identifiers (DOIs). Furthermore, they incorporate a "CITATION.cff" file, streamlining the generation of project citations in various formats, such as plain text or BibTeX.
Building React Application:
In the construction of React Applicationa, Zakodium employs a modular approach, relying on a multitude of specialized libraries for the graphical user interface (GUI), React components, and other functionalities. Impressively, over 60 libraries managed by Zakodium are integrated into NMRium, each serving specific purposes such as converters, structure viewing, and data loading and saving. A notable example is the 'nmr-load-save' library, capable of extracting comprehensive metadata from raw files within NMRium. Another case in point is 'Pixelium,' a library primarily focused on image processing. While not directly related to NMR, Pixelium extensively utilizes libraries originally created for NMRium, showcasing the versatility and cross-applicability of their development efforts. The team employs GitHub projects feature, providing a comprehensive overview of ongoing work and enabling the archiving of completed tasks. Automation is key in their workflow, with automatic bug reporting integrated into NMRium to promptly flag and address issues as they arise.
New NMRium Features
NMRium introduces a range of new features that enhance its capabilities:
- Multiplet analysis: Conduct multiplet analysis directly within the browser without the need for downloads, where you can display multiplicity trees, show the structure and do the assignment.The relation between the tables, molecule and spectra is interactive.
- Databases: NMRium incorporates a versatile database featur to have databases for solvent or references for instance. Users can perform searches based on structure, substructure, or shifts. Additionally, there is advanced search to look for specific atoms. Simulation:
- Prediction: Offering both 1D and 2D prediction capabilities, NMRium predicts spectra based on user-provided structures. It is also interactive.
- Simulation: A novel addition, the simulation feature enables spectrum generation by entering values directly into the table, which is particularly beneficial for educational purposes
- Metabolomics: NMRium excels in handling substantial gigabyte-sized files even within the browser. Filtering options enable users to navigate and extract relevant information from large amount of files based on specific criteria such as the type.
- e-learning: You process the spectrum and and propose a structure and you find if correct or not.
Formats Discussion
In our considerations of data formats, it's crucial to draw a distinction between archival formats and those tailored for processing. A case in point is mzML, which, despite its stability over time, has faced reservations among users for certain applications. In Massbank, there exists a diversity of formats, including the less-documented Bruker MS, in contrast to the well-documented Bruker NMR. In the context of NMRium, an approach encapsulating both original and processed data within a single JSON file is adopted. Notably, the utilization of JSON holds the promise of longevity, as its readability is anticipated to endure over the next 30 years. Audit trails are needed and we need to be able to read the data in 30 years from now.
Towards Open NMR Data Formats - What We Want and What We Can Have
Dr. Johannes Liermann Presentation
We prepare a sample with a molecule and in the spectrometer we get information that we want to assign to the molecule, which results in several layers of NMR data: raw data, processed data, assignment data and another layer that is the metadata. In higher levels in NMR it is difficult to tell what is data and what is metadata.
There are several NMR formats available:
- JCAMP: Text-based format with tags defining both metadata and actual data along with the binary data, making it lengthy if uncompressed.
- Bruker: Utilizes a folder structure with various files. 'fid' file holds binary data from acquisition. 'acqu' file relates to JCAMP and provides metadata. Bruker dataset is comprehensive, containing all necessary elements to reproduce an NMR experiment.
- nmrML: Employs an embedded ontology, distinguishing itself as probably the only one of the NMR formats utilizing ontology. Predominantly recognized in the metabolomics and biology domains but can be adapted for broader use.
- NMReData: Focuses beyond spectral data, extending into sdf structure and enriching it with 1D, 2D, assignments, coupling constants, and correlations.
The landscape: JCAMP and Bruker cover raw and processed data and if at all assignment data. nmrML covers a little assignment but not designed for that. NMReDara is covers only assignment data.
Challenges Examples:
- Flip angle in Bruker: Obtaining the flip angle information requires reference to the pulse sequence, which may be clear for chemists but poses difficulty for developers. Recognizing the specific NMR experiment, such as HMBC, within a repository can indeed be challenging due to the diverse pulse sequence names. However, they all follow a machine readable syntax that can identify the experiment.
- TOCSY Experiment Naming Complexity: TOCSY experiment names can be intricate, especially when the term "TOCSY" is not explicitly mentioned.
- Deciphering Pulse Program Information: In cases like d8 representing the mixing time, finding the relevant information in the dataset file (D field) may be challenging as you have to find the 8th one.
- Locating Coupling Constants: Coupling constants may be embedded in the pulse sequence, requiring additional steps for retrieval.
- Ontologies: Maintaining consistency in units, solvent values, and experiment types can be challenging without standardized ontologies. We need to actively contribute to and utilize available NMR ontologies
Discussion:
Establishing a mapping between ontology descriptions and vendors' datasets emerges as a crucial step. Later on, software can take an ontology and select a relevant branch of it for example to ask the user to pick a solvent. To initiate the effective integration of ontologies into a format, the we need following considerations: The primary requirement is to incorporate unique identifiers for ontology terms, as the software doesn’t need to understand the semantic of the term at this level. Integration of a unit ontology can enhance the consistency and accuracy of data representation.
What do we want from an open NMR format? Do we need one to start with?
-
Bruker's documentation being open is a positive step. If it becomes even more widely adopted, it could potentially fulfill the needs of the NMR community, and it might reduce the immediate need for a separate open format. Developing and maintaining an open format requires considerable resources and collaboration. Modernizing Bruker formats might be a pragmatic approach, given their widespread usage.
-
JCAMP's popularity for Electronic Lab Notebooks (ELNs) makes it challenging to replace with Bruker formats,. Overlapping documentation for various analytical techniques using JCAMP can lead to confusion and hinder effective utilization. Despite JCAMP's capacity for metadata, there are limitations in including crucial information, emphasizing the need for enhancements. JCAMP's textual nature poses challenges for modern parsers, discouraging efforts to update and modernize it. The lack of a standardized way to validate JCAMP, requiring parsers writing for all "standard" versions, adds complexity to the format. Introducing ontologies into JCAMP may face resistance due to concerns that such modifications might not align with the preferences or understanding of chemists.
Collaborating with IUPAC by incorporating elements from Bruker and seeking IUPAC recommendations could be a strategic approach to improving and modernizing JCAMP. We can enhance JCAMP by identifying and determining the minimum information missing there and incorporating them. Utilizing the dot in JCAMP for optional fields could offer a structured way to add relevant information specific to NMR.
We Are Still Stuck With The Question of What Format to Go With
- The consensus is to focus on enhancing existing formats rather than developing entirely new ones. The applied modifications should remain minimal.
- Acknowledgment that JCAMP, while popular, has limitations and misses important elements. This serves as a motivation to enhance and improve the format to capture more comprehensive data. A suggestion to take the JCAMP model and provide an additional serialization in JSON format, while emphasizing the importance of validation and testing, particularly for industry use.
Conclusion:
- Considering the option of having the existing files with an additional JSON file for metadata. We need use cases where available formats fall short is highlighted. Existing NMR formats are not perceived as "bad enough" to warrant the development of entirely new formats. This reflects a pragmatic approach to evolving the current standards.
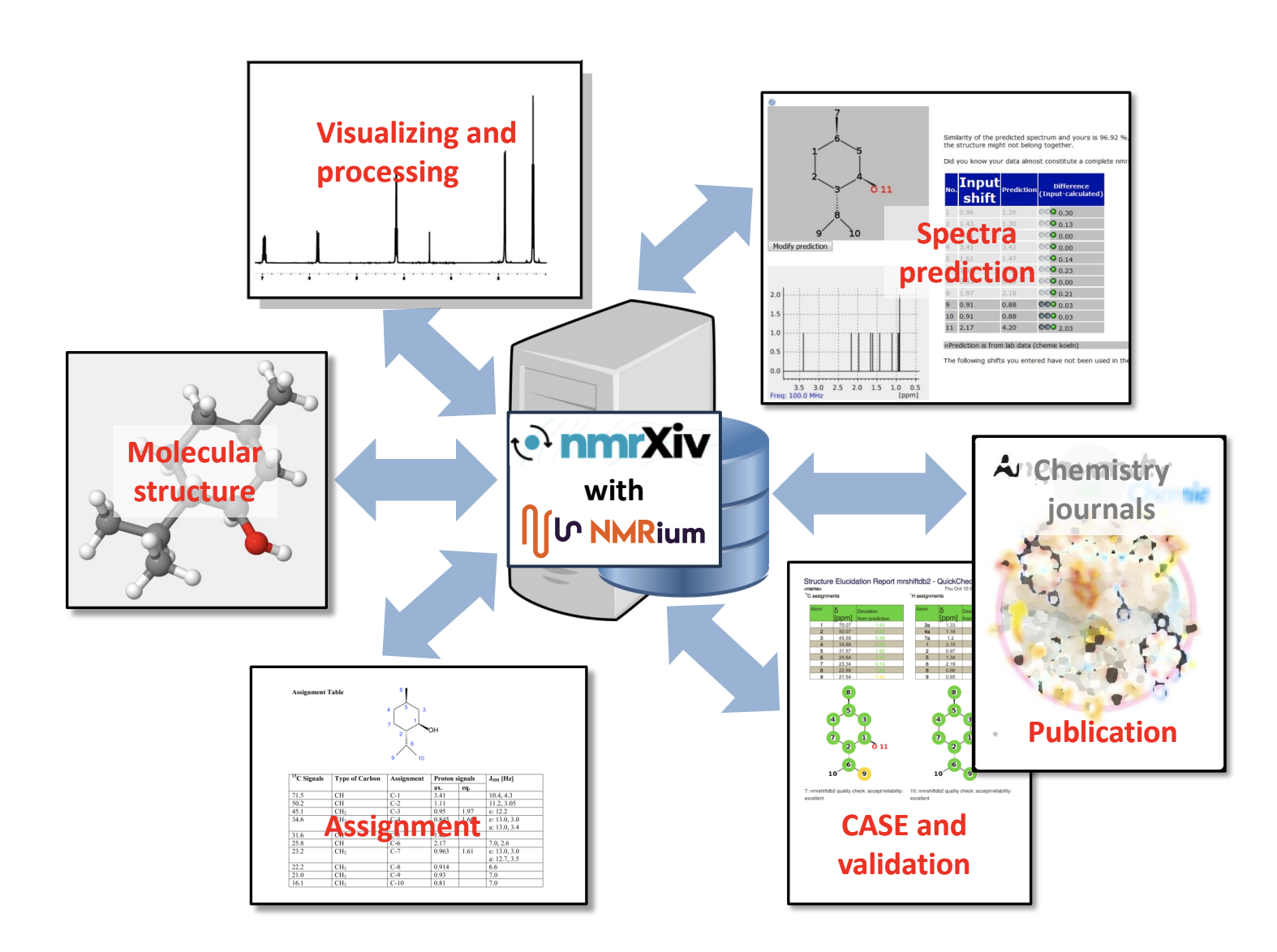
nmrXiv + NMRium as The Open Source Choice for NMR Platforms
Dr. Nils Schlörer Presentation
The focus is on academia, not the industry. What tasks in the lab and the ideal workflow and what bottlenecks we have. What features we could have by merging nmrium with nmrXiv in such environment. Laboratories lack standardization, with variations in tools, systems, and practices, including the use of various systems such as LIMS (Laboratory Information Management Systems) and servers. Exploring the potential benefits of open access and automation in academic labs, with a particular emphasis on facilitating data submission without the need for a formal paper submission process, and providing the data to the user with accounts.
The repository, especially when integrated with applications like NMRium, becomes a valuable resource not only for visualization but also for data processing and assignment. Repositories have two aspects:
- Administrator's Role: Involves quality checks, ranking, and overall management of the repository.
- User Expectations: Consideration of user expectations and the importance of user-friendly interfaces. Understanding user needs is crucial, and overly complex or needy submission processes may deter users.
Highlighting the importance of providing added value to users. For example, NMRium suggesting assignments for 2D spectra, which is a common need in routine analyses.
Ideal Laboratory NMR Workflow
The ideal laboratory NMR workflow for generating data ready for submission involves several key considerations to streamline the process and enhance user experience:
- Hosted Data Storage: Encourage the hosting of data in repositories rather than relying on local storage. Overcoming the preference for local storage by chemists can enhance data accessibility and sharing.
- Structure Linkage: Ensure that each spectrum is linked to at least one corresponding chemical structure. Despite being common, the practice of not submitting structures should be hindered
- Consistent Local and Public Format: Promote the use of a consistent format for both local and public data storage. This minimizes the need for users to switch between different systems, ensuring a seamless workflow.
- Integrated Visualization and Processing: Provide tools for direct visualization and processing of data on the same platform after data is created. Integrated capabilities eliminate the need for users to rely on external software, enhancing efficiency.
- Living Data with NMRium: Implement the concept of "living data" with tools like NMRium in platforms such as nmrXiv. This approach ensures that data remains dynamic and interactive, allowing users to explore and analyze it comprehensively.
- Digital Availability of 2D Information: Shift from the current practice where people distribute some digital data and do paper assignment and the 2D information is usually mainly ignored.
- Unified Interface for All Tasks: Provide a unified interface that caters to all tasks, from data submission to visualization and processing.
- Suggested Assignment Feature: Incorporate a feature that suggests assignments to users. This not only adds value, but also makes the platform more attractive and user-friendly.
- Assignment Control: There should be some assignment control to check the plausibility by chemical shift comparisons but also can be used with case environment including logic, and if people do some assignment the system should be capable of continuing the job.
Success Factors:
The success factors for an ideal NMR repository revolve around simplifying the user experience. Here's a breakdown of the key success factors:
-
Minimizing Interfaces: Chemists are averse to handling multiple interfaces, potentially leading them to revert to traditional paper methods. The repository should provide a unified interface, streamlining all steps of the NMR workflow featured below:
-
Consistent Structure Provision: Ensuring that structures are consistently provided with every NMR dataset submission.
-
Privacy and Data Sharing Options: Privacy concerns users and we need flexible data sharing options, offering robust privacy features, including the ability to publish data with restricted access to specific groups. Providing users with options for controlling data visibility fosters trust and compliance instead of just missing the data.

Discussion:
The discussion underscores the importance of including Electronic Lab Notebooks (ELNs) in the vision for NMR data management. Core facilities should play the role of raising the bar for data digitization. However, not all facilities may currently have the autonomy to enforce these practices, but they still should try for the better. Funders, such as DFG, can influence data quality by tying funding to specific standards. This creates a mechanism for control over data quality from the funder's perspective. The use of commercial management systems in the Jena platform lead to replacing traditional paper-based workflows. This shift underscores the practical adoption of digital tools. However, searching is based on people and groups rather than molecules. This limitation is attributed to the non-mandatory submission of structures.
nmrXiv repository layer is ready, with ongoing enhancements to the submission and molecule layers. Plans include building prediction and search layers separate from nmrXiv to facilitate broader usage. We already work with Chemotion, an open-source ELN that we found to be the best in the chemistry domain.
NMR Data Packaging with Metadata in NFDI4Chem
NMR Metadata Availability in NFDI4Chem: NFDI4Chem services, including nmrXiv (NMR data repository) and Chemotion (Repository for samples, reactions, and the accompanying analytical data), provide NMR metadata in a structured JSON format. This format adheres to schema.org and Bioschemas types, ensuring standardized representation. The use of NMRium in nmrXiv and Chemotion ensures a high extraction of metadata. This metadata can be translated into schema JSON and further optimized to include all the minimum information, while recommended and optional data can still partially depends on user's input. The platforms already provide the means for users to input some of this data.
Metadata Loss during Data Download: Currently, for both repositories, when users download data, no structured metadata files are automatically added within the folder. This lack of inclusion poses a risk of potential metadata loss during the download process. In Chemotion, even datasets belonging to the same reaction need to be downloaded individually. This can be cumbersome and may contribute to a fragmented dataset. Users need to go through a separate download/export process to obtain metadata. This involves using separate download buttons (Chemotion) or making API calls (nmrXiv), adding an extra step to access metadata.
In Chemotion, it is common to find additional chemical materials in the "description" field that may not be present in the reaction table. This adds complexity when deciding what metadata to export, as there may be supplementary information in unstructured fields.
The need to package related data and metadata together in one folder is crucial for efficient data management. Data packaging involves using tools to containerize data along with its associated metadata in a format that is both human- and machine-readable.
Data packaging tools request metadata to be structured, ensuring that it follows a standardized format that is both human- and machine-readable. Some tools incorporate data integrity checks to monitor the data and ensure its consistency throughout different processes, such as downloading, zipping/unzipping, and transferring. The tools also provide packaged data in formats that are convenient to transfer and download.
Data Packaging Approaches
BagIt
BagIt provides file layout conventions for data packaging. Here's an overview of the key components of BagIt:
- Root Folder - "Bag": The root folder, referred to as the "bag," is the primary container for organizing the data. It is has two types of files:
- Payload Files: The main data files.
- Tag Files: Tag files serve a descriptive role, providing metadata or additional information about the contents of the "bag."
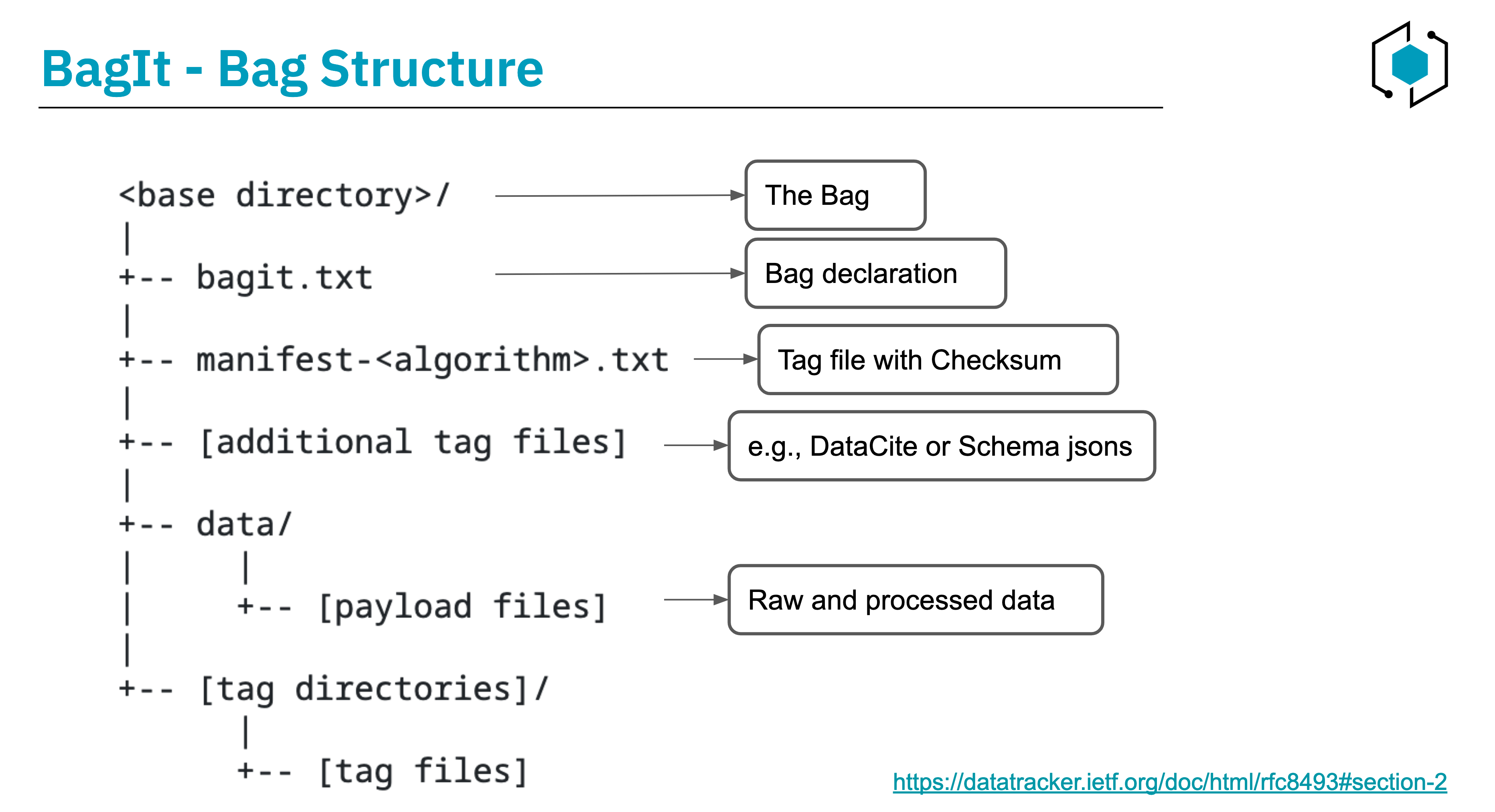
The BagIt structure provides a clear and organized approach for data packaging. Here's a summary of the key components and requirements of the bag (root folder):
- Bag Declaration (bagit.txt): The "bagit.txt" tag file establishes the BagIt version and character encoding for tag files. It MUST consist of exactly two lines in this order:
- BagIt-Version: M.N
- Tag-File-Character-Encoding: ENCODING
- Payload Manifest (manifest-algorithm.txt): The payload manifest file lists each payload file name alongside a corresponding checksum. This allows for robust data integrity checking. A bag can have multiple payload manifests, each using a different checksum algorithm.
- Additional Tag Files: BagIt allows for the inclusion of other tag files. For instance, adding DataCite metadata or RO-Crate JSON files enhances the bag's descriptive capabilities and interoperability with different metadata standards.
- Subdirectory "data": The base directory MUST contain a subdirectory named "data."
BagIt has its format to integrate checksum algorithms allowing data integrity checking. It additionally offers flexibility in the folder structure to add different metadata files as it suits the user. However, it doesn’t provide a structured way NMR related metadata. And even its structured specifications are txt based (https://datatracker.ietf.org/doc/html/rfc8493#section-2.2.2 )
RO-Crate
RO-Crate (Research Object Crate) is a method of aggregating and describing research data with associated metadata. This includes information about its creators, the equipment utilized, and the licensing terms for subsequent reuse.
RO-Crate leverages JSON-LD (JSON for Linked Data) to express metadata, describing data resources as well as contextual entities such as people, organizations, software, and equipment as a series of linked JSON-LD objects, utilizing commonly adopted vocabularies, primarily schema.org.
- The root directory is identifiable by the presence of the RO-Crate Metadata File, named ro-crate-metadata.json.
- The core of RO-Crate is a JSON-LD file, the RO-Crate Metadata File, named ro-crate-metadata.json. This file contains structured metadata about the dataset as a whole (the Root Data Entity). Optionally, this file may also contain metadata about some or all of the individual files within the dataset. This structured metadata file offers a straightforward means to assert assert the authors (e.g. people, organizations) of the RO-Crate or one its files. Additionally, it enables the capture of more intricate provenance information for files, encompassing details about how they were created using specific software and equipment.
- Payload files and directories: These are the actual files and directories that make up the dataset being described.
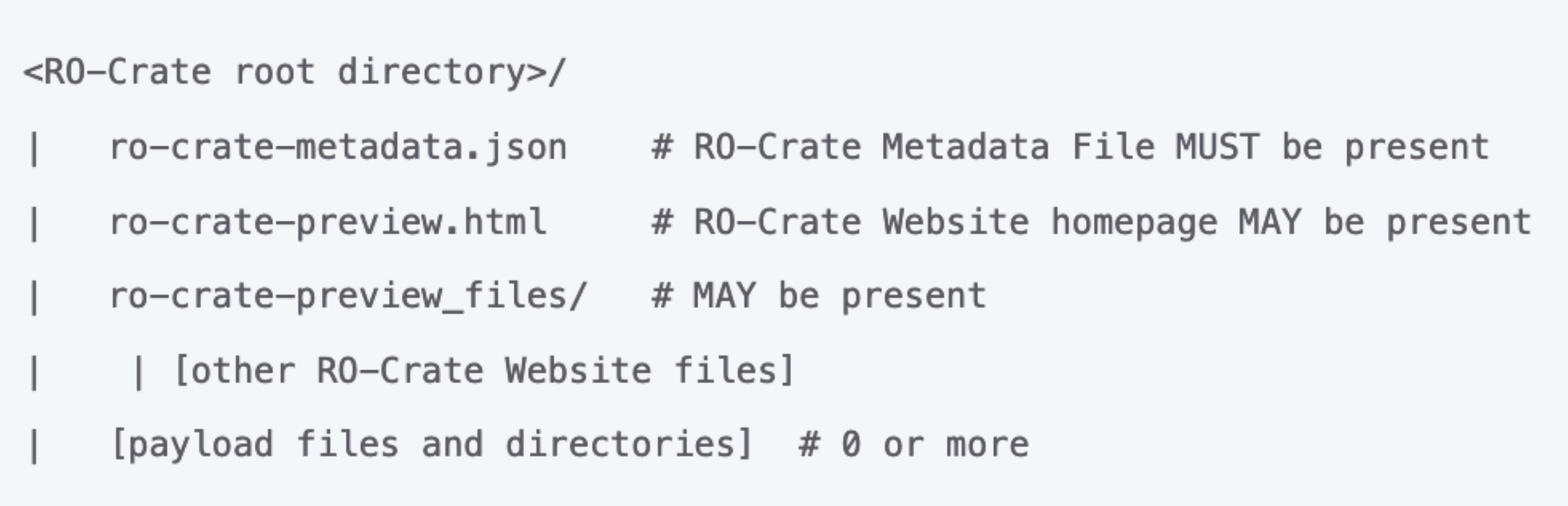
An important distinction exists between the interpretation of datasets in RO-Crate and nmrXiv from NFDI4Chem, particularly in the categorization of folders within the root directory. While RO-Crate designates all folders within any level of the root as "Datasets," nmrXiv considers a complete NMR assay (e.g., a Bruker folder or jcamp file) as a dataset, causing a huge confusion. This discrepancy becomes evident when, for example, a "Bruker" folder is present within the root, qualifying as a dataset according to RO-Crate's definition. Simultaneously, a "pdata" folder within the Bruker folder may also be considered a dataset within the RO-Crate framework. In contrast, nmrXiv views the entire assay folder as a dataset.
Additionally, two identical Bruker and Jcamp assays will have different types in RO-Crate. The presentation demos many scenarios which won't work, including representing The NMR assay as RO-Crate root or grouping them under the root.
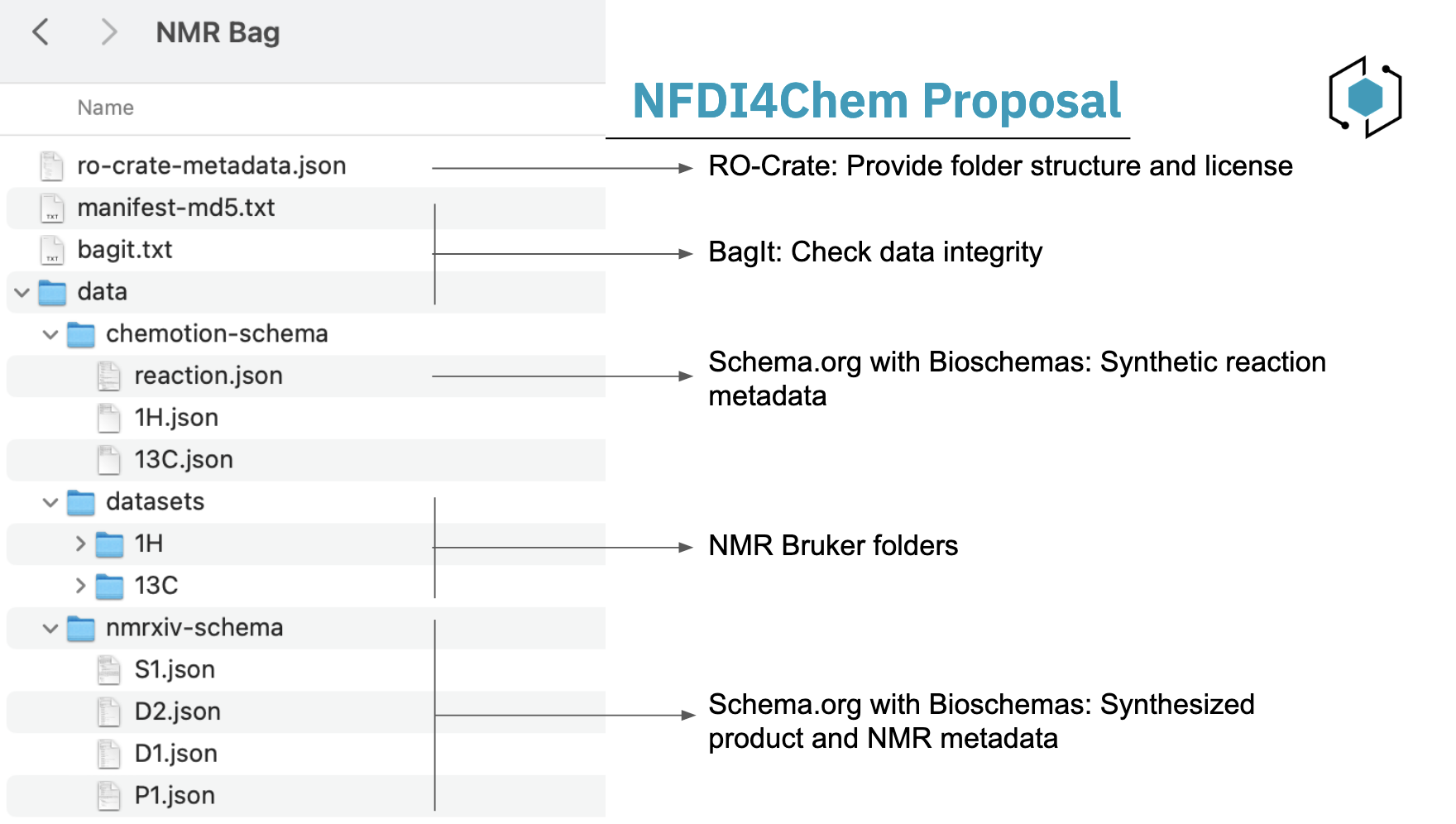
NFDI4Chem Proposal
NFDI4Chem advocates for a synergistic approach that harnesses the strengths of multiple packaging tools along with schema.org metadata to enhance the representation of NMR-specific data. The proposed combination includes:
- BagIt: For data integrity check.
- RO-Crate: For folders structure.
- Schema.org: For NMR-specific metadata.
Here is a link to an exemplary NMR package.
nmrCV Controlled Vocabulary
nmrCV, was initially developed as part of nmrML before 2019, and we still use it as an NMR controlled vocabulary. nmrCV still needs:
- Structure cleaning in terms of hierarchy.
- Identifying and addressing missing terms.
- Collaboration and importing relevant content.
- Using ODK to dockerize the CV and manage its continuous integration.